data(voomed_GBM, package="plotlyvignettes")
data(GBMdata, package="plotlyvignettes")
pcs <- prcomp(t(voomed_GBM$E))
pc_data <- pcs$x
columns <- c(
"paper_IDH.status",
"paper_Age..years.at.diagnosis.",
"paper_Gender",
"paper_Pan.Glioma.RNA.Expression.Cluster",
"ethnicity"
)
colours <- colData(GBMdata)[, columns, drop = FALSE]
colours <- as.data.frame(colours)
colours$TotalReads <- colSums(assay(GBMdata))
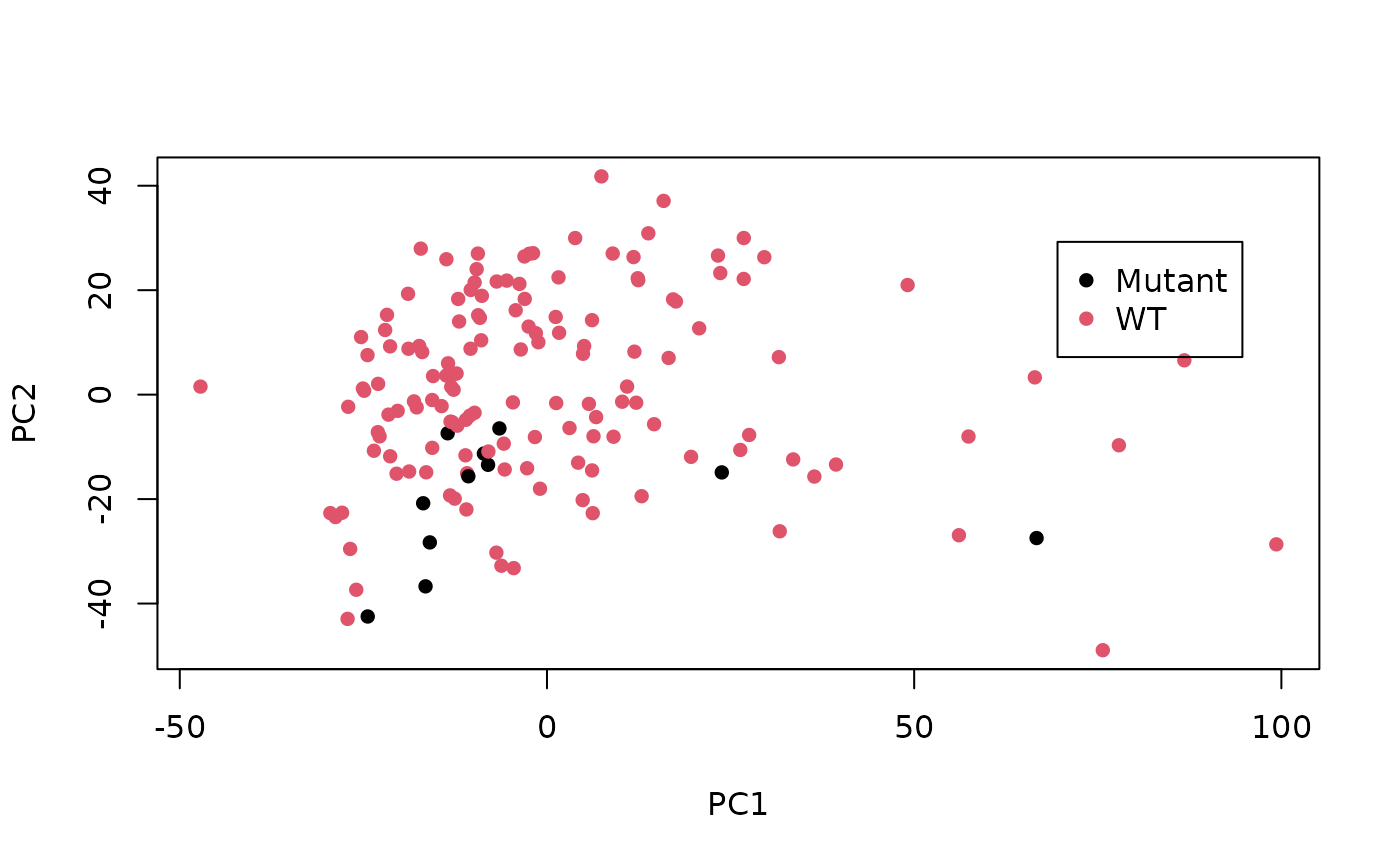
plot(
pc_data[, 1],
pc_data[, 2],
col = as.factor(colours[[1]]),
pch = 16,
xlab = "PC1",
ylab = "PC2")
legend(
max(pc_data[, 1]) * 0.7,
max(pc_data[, 2]) * 0.7,
unique(colours[[1]]),
pch = 16,
col = unique(colours[[1]]))